Introduction to Data Visualization for Meta-Analysis
with tidymeta and ggplot2
Malcolm Barrett
Install R: bit.ly/pm605_r
Handout: bit.ly/pm605_tut
04/23/2018
1 / 73
Data Visualization with R
ggplot2 and the tidyverse are friendly and consistent tools for data analysis and visualization
2 / 73
Data Visualization with R
ggplot2 and the tidyverse are friendly and consistent tools for data analysis and visualization
Better plots are better communication
3 / 73
Data Visualization with R
ggplot2 and the tidyverse are friendly and consistent tools for data analysis and visualization
Better plots are better communication
tidymeta makes it easy to manipulate and plot meta-analysis results
4 / 73
Introduction to the Data
What's the impact of intrauterine device (IUD) use on risk of cervical cancer?
5 / 73
Introduction to the Data
What's the impact of intrauterine device (IUD) use on risk of cervical cancer?
16 studies: 4,945 cases and 7,537 controls
6 / 73
Introduction to the Data
What's the impact of intrauterine device (IUD) use on risk of cervical cancer?
16 studies: 4,945 cases and 7,537 controls
Women who used IUDs were at a third less risk than those who didn't (OR 0.64)
7 / 73
library(tidymeta)iud_cxca## # A tibble: 16 x 26## study_id study_name author es l95 u95 lnes lnl95 lnu95## <int> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>## 1 1 Roura, 2016 Roura 0.600 0.300 1.20 -0.511 -1.20 0.182 ## 2 2 Lassise, 1991 Lassi… 0.800 0.500 1.20 -0.223 -0.693 0.182 ## 3 3 Li, 2000 Li 0.890 0.730 1.08 -0.117 -0.315 0.0770## 4 4 Shields, 2004 Shiel… 0.500 0.300 0.820 -0.693 -1.20 -0.198 ## 5 5 Castellsague… Caste… 0.630 0.380 1.06 -0.462 -0.968 0.0583## 6 6 Castellsague… Caste… 0.450 0.300 0.670 -0.799 -1.20 -0.400 ## 7 7 Brinton, 1990 Brint… 0.690 0.500 0.900 -0.371 -0.693 -0.105 ## 8 8 Parazzini, 1… Paraz… 0.600 0.300 1.10 -0.511 -1.20 0.0953## 9 9 Williams, 19… Willi… 1.00 0.600 1.60 0. -0.511 0.470 ## 10 10 Hammouda, 20… Hammo… 0.300 0.100 0.500 -1.20 -2.30 -0.693 ## 11 11 Castellsague… Caste… 1.08 0.370 3.20 0.0770 -0.994 1.16 ## 12 12 Castellsague… Caste… 0.340 0.0500 2.56 -1.08 -3.00 0.940 ## 13 13 Castellsague… Caste… 0.870 0.340 2.23 -0.139 -1.08 0.802 ## 14 14 Castellsague… Caste… 0.490 0.190 1.23 -0.713 -1.66 0.207 ## 15 15 Castellsague… Caste… 0.240 0.0900 0.660 -1.43 -2.41 -0.416 ## 16 16 Celentano, 1… Celen… 0.500 0.170 1.47 -0.693 -1.77 0.385 ## # ... with 17 more variables: selnes <dbl>, group <fct>, case_num <dbl>,## # control_num <dbl>, start_recruit <dbl>, stop_recruit <dbl>,## # pub_year <dbl>, numpap <dbl>, ses <dbl>, gravidity <dbl>,## # lifetimepart <dbl>, coitarche <dbl>, hpvstatus <dbl>, smoking <dbl>,## # location <chr>, aair <dbl>, hpvrate <dbl>8 / 73
Five variables from iud_cxca we'll use
study_name
lnes
selnes
group
pub_year
9 / 73
Five variables from iud_cxca we'll use
study_name = Author + study year
lnes
selnes
group
pub_year
10 / 73
Five variables from iud_cxca we'll use
study_name
lnes = ln(Odds Ratio)
selnes
group
pub_year
11 / 73
Five variables from iud_cxca we'll use
study_name
lnes
selnes = SE of ln(OR)
group
pub_year
12 / 73
Five variables from iud_cxca we'll use
study_name
lnes
selnes
group = Study design
pub_year
13 / 73
Five variables from iud_cxca we'll use
study_name
lnes
selnes
group
pub_year = Publication year
14 / 73
Meta-Analysis Plot Types
15 / 73
Meta-Analysis Plot Types
Forest Plot
16 / 73
forest_plot()
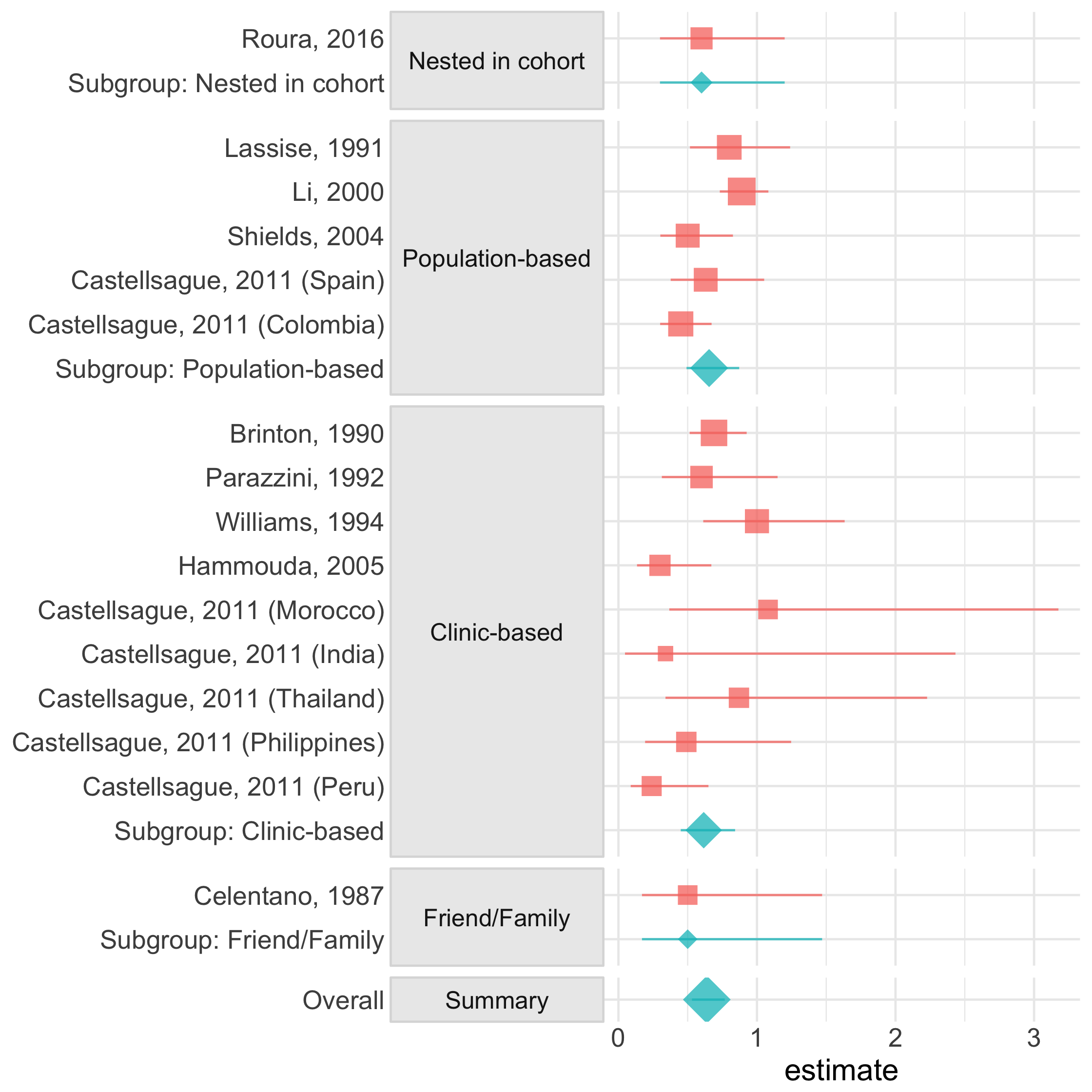
17 / 73
Meta-Analysis Plot Types
Forest Plot
Funnel Plot
18 / 73
funnel_plot()
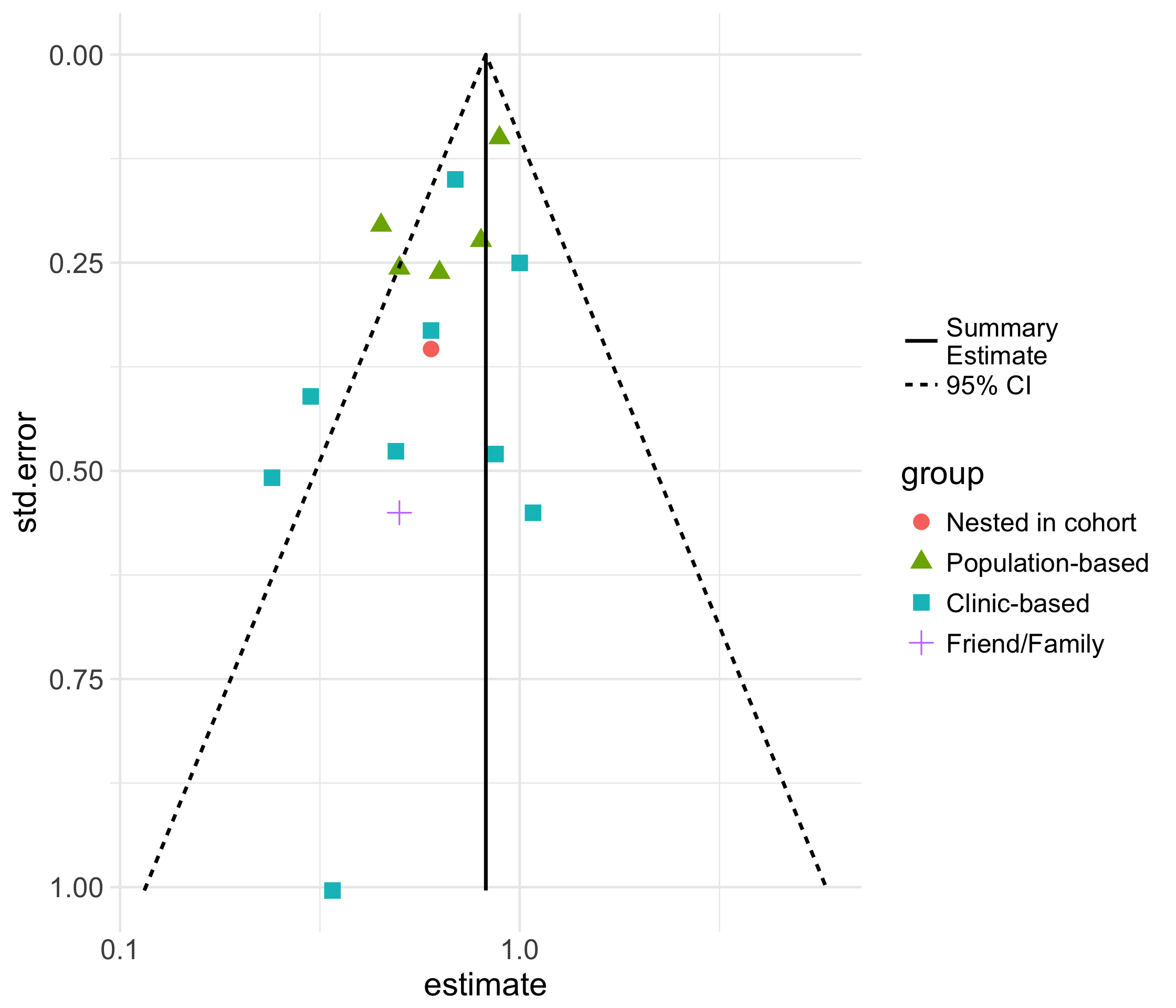
19 / 73
Meta-Analysis Plot Types
Forest Plot
Funnel Plot
Influence/Sensitivity Plot
20 / 73
influence_plot()
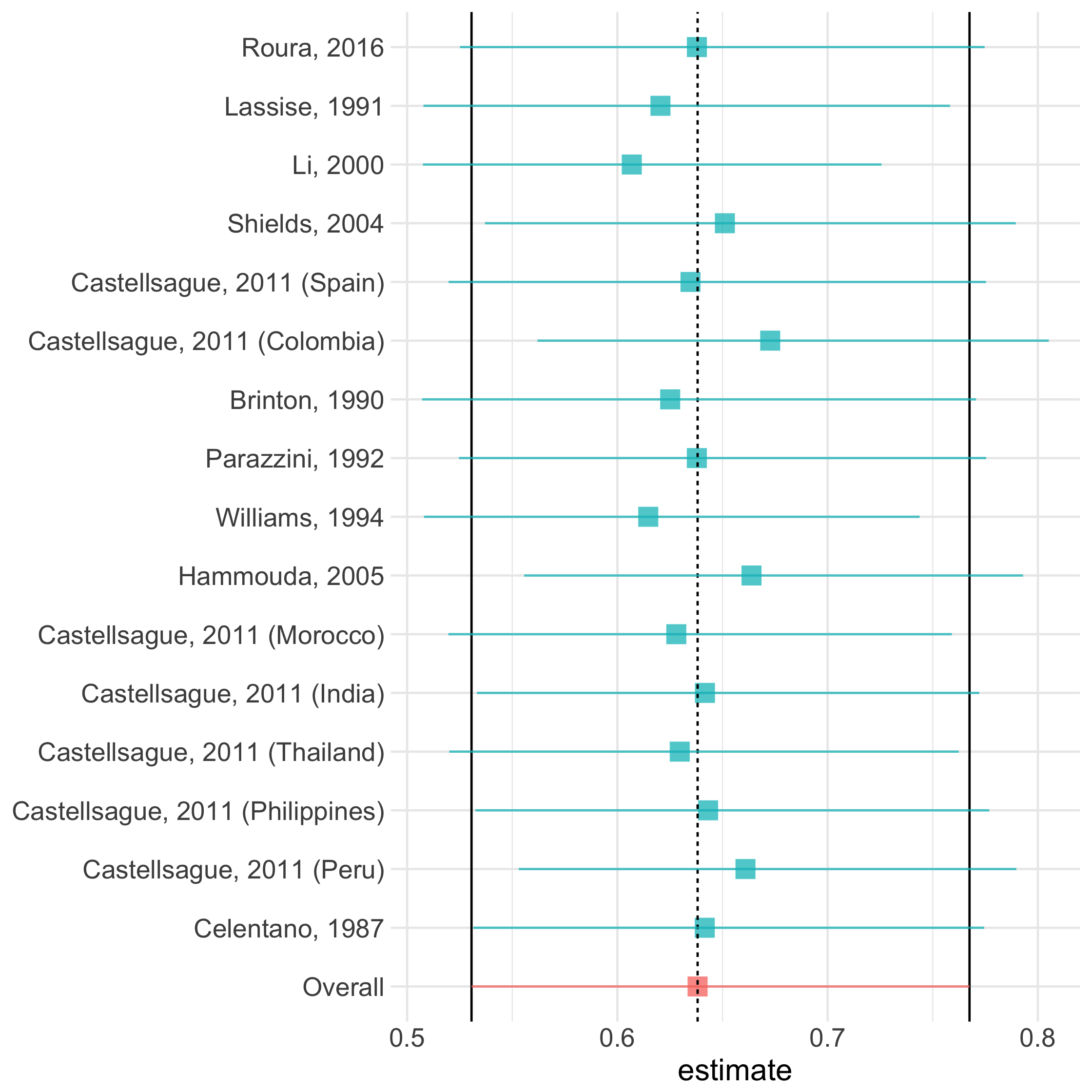
21 / 73
Meta-Analysis Plot Types
Forest Plot
Funnel Plot
Influence/Sensitivity Plot
Cumulative Plot
22 / 73
cumulative_plot()
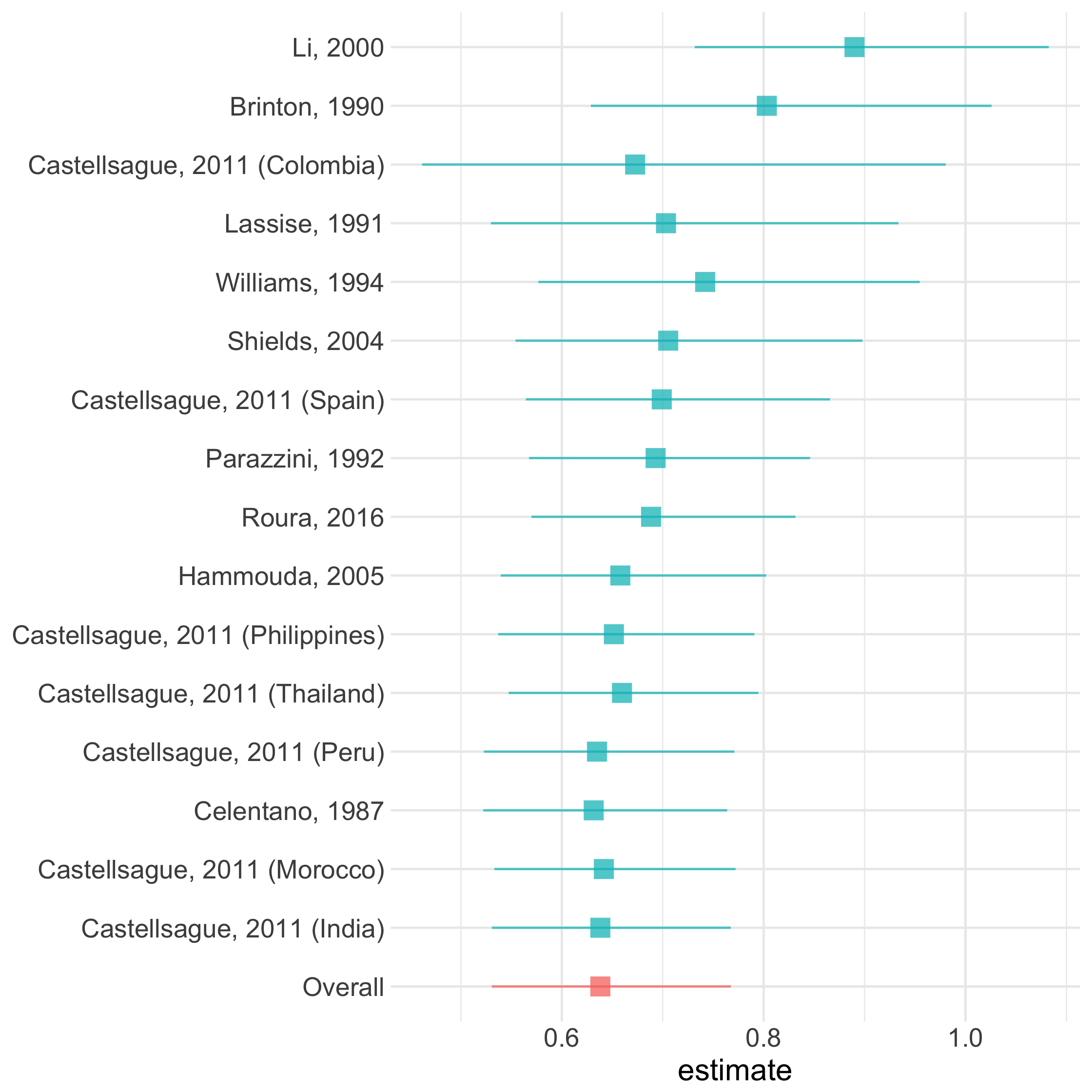
23 / 73
A Crash Course in the Tidyverse
24 / 73
ggplot2: Elegant Data Visualizations in R
25 / 73
ggplot2: Elegant Data Visualizations in R
Based on a Grammar of Graphics
26 / 73
ggplot2: Elegant Data Visualizations in R
Based on a Grammar of Graphics
Data is mapped to aesthetics; Statistics and plot are linked
27 / 73
ggplot2: Elegant Data Visualizations in R
Based on a Grammar of Graphics
Data is mapped to aesthetics; Statistics and plot are linked
Sensible defaults; Infinitely extensible
28 / 73
library(ggplot2)p <- ggplot(iud_cxca, aes(case_num + control_num, lnes, color = group))p
29 / 73
library(ggplot2)p <- p + geom_point()p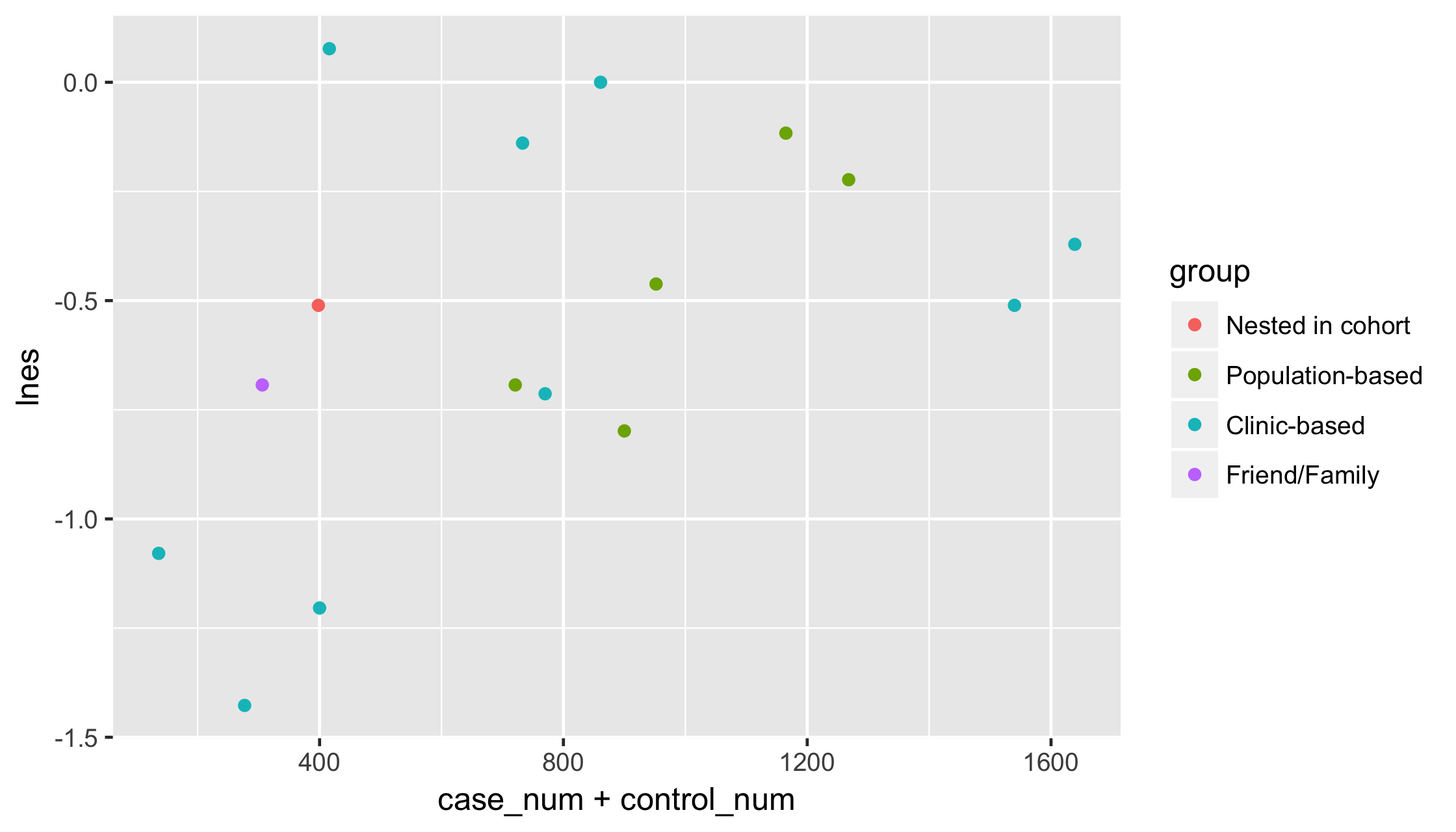
30 / 73
p <- p + geom_smooth(method = "lm", se = FALSE)p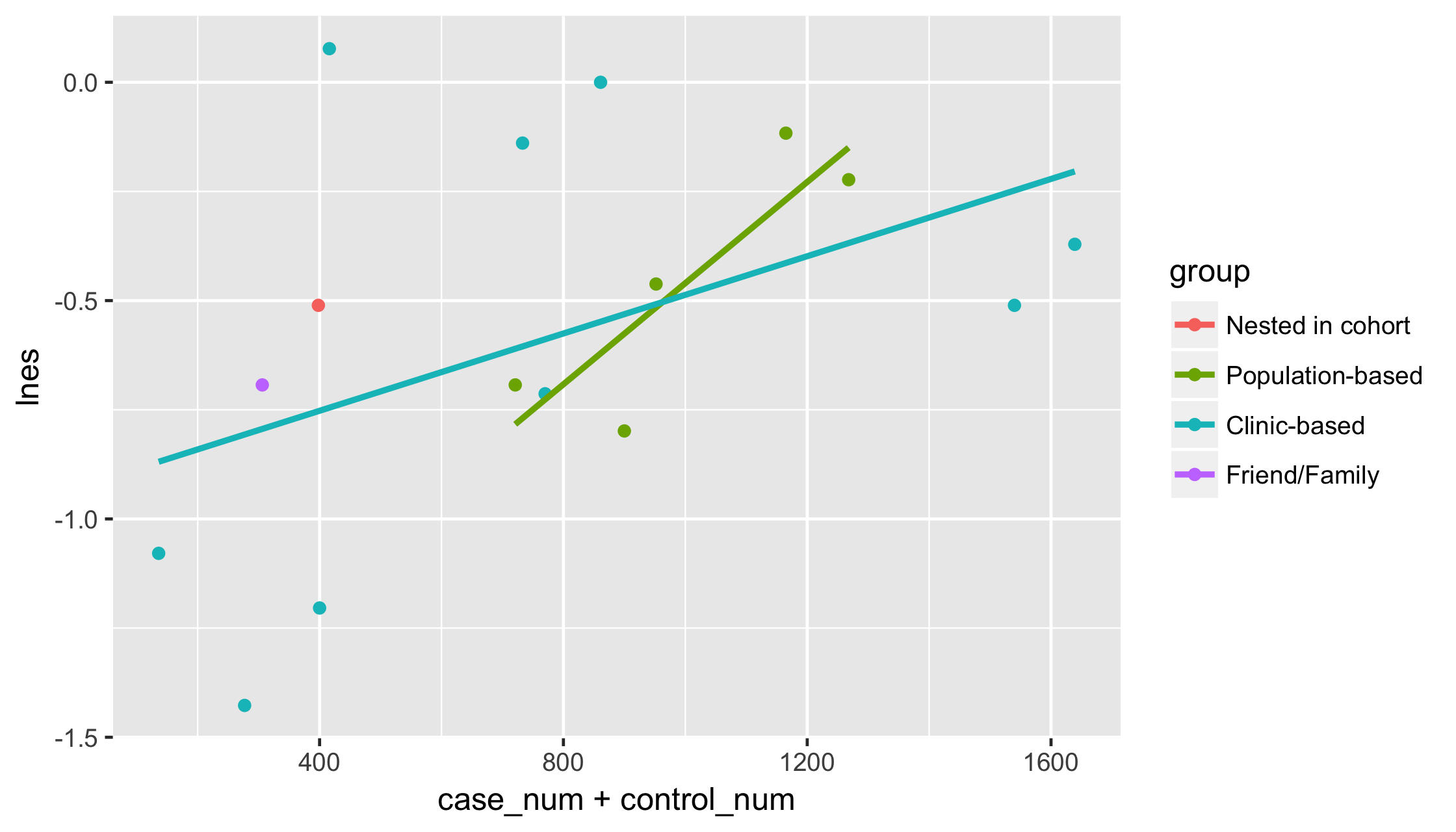
31 / 73
p + labs(title = "The Effect of Sample Size on Estimate", x = "Sample Size", y = "ln(Odds Ratio)") + scale_color_discrete(name = "Study Design") + theme_minimal() + theme(text = element_text(size = 16))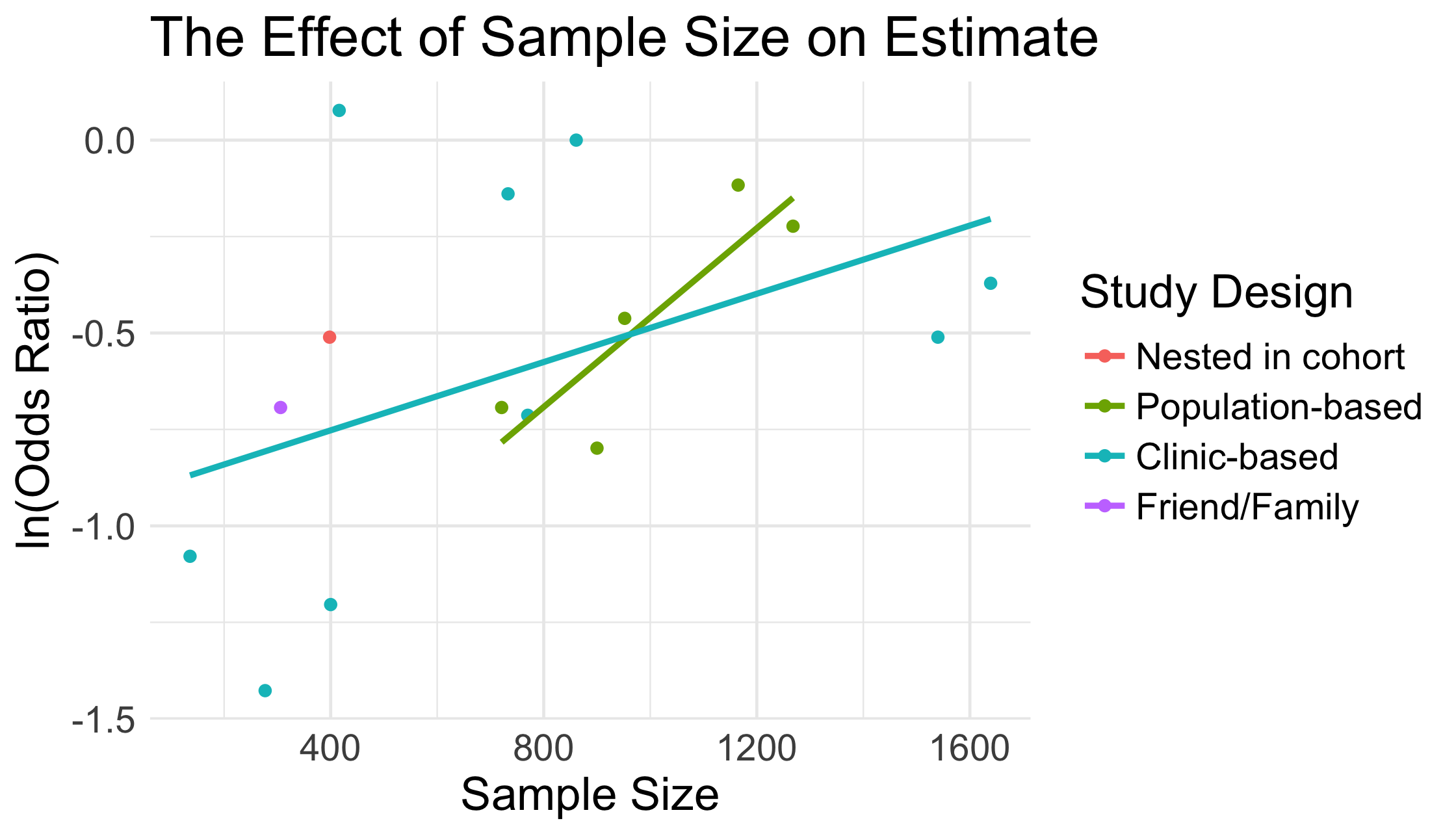
32 / 73
Tidy Data is Easier to Plot

33 / 73
Tidy Data is Easier to Plot

Each column is a single variable
34 / 73
Tidy Data is Easier to Plot

Each column is a single variable
Each row is a single observation
35 / 73
Tidy Data is Easier to Plot

Each column is a single variable
Each row is a single observation
Each cell is a value
36 / 73
Our Tidy Tools
%>%
mutate()
arrange()
group_by()
tidy()
37 / 73
Our Tidy Tools
%>%: passes the results of one function to the next
mutate()
arrange()
group_by()
tidy()

38 / 73
Our Tidy Tools
%>%
mutate(): changes or creates a new variable
arrange()
group_by()
tidy()

39 / 73
Our Tidy Tools
%>%
mutate()
arrange(): sorts a data set by a variable
group_by()
tidy()

40 / 73
Our Tidy Tools
%>%
mutate()
arrange()
group_by(): groups a data set by a variable
tidy()

41 / 73
Our Tidy Tools
%>%
mutate()
arrange()
group_by()
tidy(): tidies statistical results

42 / 73
Tidy Meta-Analysis
meta_analysis()
43 / 73
Tidy Meta-Analysis
meta_analysis()
ma <- iud_cxca %>% group_by(group) %>% meta_analysis(yi = lnes, sei = selnes, slab = study_name, exponentiate = TRUE)ma## # A tibble: 21 x 11## group study type estimate std.error statistic p.value conf.low## <fct> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>## 1 Nested … Roura, 2… study 0.600 0.354 -1.44 NA 0.300## 2 Nested … Subgroup… summ… 0.600 0.354 -1.44 0.149 0.300## 3 Populat… Lassise,… study 0.800 0.223 -0.999 NA 0.516## 4 Populat… Li, 2000 study 0.890 0.0999 -1.17 NA 0.732## 5 Populat… Shields,… study 0.500 0.257 -2.70 NA 0.302## 6 Populat… Castells… study 0.630 0.262 -1.77 NA 0.377## 7 Populat… Castells… study 0.450 0.205 -3.90 NA 0.301## 8 Populat… Subgroup… summ… 0.655 0.146 -2.90 0.00374 0.492## 9 Clinic-… Brinton,… study 0.690 0.150 -2.47 NA 0.514## 10 Clinic-… Parazzin… study 0.600 0.331 -1.54 NA 0.313## # ... with 11 more rows, and 3 more variables: conf.high <dbl>,## # meta <list>, weight <dbl>44 / 73
Forest Plot
forest_plot()
45 / 73
Forest Plot
forest_plot()
ma %>% forest_plot(group = group)46 / 73
Forest Plot
forest_plot()
ma %>% forest_plot(group = group)text_table()
47 / 73
Forest Plot
forest_plot()
ma %>% forest_plot(group = group)text_table()
ma %>% text_table(group = group, "Weights" = weight)48 / 73
patchwork: Compose ggplots

49 / 73
patchwork: Compose ggplots
Join ggplots quickly and accurately

50 / 73
patchwork: Compose ggplots
Join ggplots quickly and accurately
library(patchwork)forest_plot() + text_table()
51 / 73
Funnel Plot
funnel_plot()
52 / 73
Funnel Plot
funnel_plot()
ma %>% funnel_plot(log_summary = TRUE)53 / 73
Influence Plot
sensitivity()
54 / 73
Influence Plot
sensitivity()
ma %>% sensitivity(exponentiate = TRUE)55 / 73
Influence Plot
sensitivity()
ma %>% sensitivity(exponentiate = TRUE)influence_plot()
56 / 73
Influence Plot
sensitivity()
ma %>% sensitivity(exponentiate = TRUE)influence_plot()
ma %>% sensitivity(exponentiate = TRUE) %>% influence_plot()57 / 73
Cumulative Plot
cumulative()
58 / 73
Cumulative Plot
cumulative()
ma %>% arrange(desc(weight)) %>% cumulative(exponentiate = TRUE)59 / 73
Cumulative Plot
cumulative()
ma %>% arrange(desc(weight)) %>% cumulative(exponentiate = TRUE)cumulative_plot()
60 / 73
Cumulative Plot
cumulative()
ma %>% arrange(desc(weight)) %>% cumulative(exponentiate = TRUE)cumulative_plot()
ma %>% arrange(desc(weight)) %>% cumulative(exponentiate = TRUE) %>% cumulative_plot(sum_lines = FALSE)61 / 73
Importing Stata data, saving ggplots
62 / 73
Importing Stata data, saving ggplots
haven: read_dta()
63 / 73
Importing Stata data, saving ggplots
haven: read_dta()
library(haven)data <- read_dta("stata_data.dta")64 / 73
Importing Stata data, saving ggplots
haven: read_dta()
library(haven)data <- read_dta("stata_data.dta")ggplot2: ggsave()
65 / 73
Importing Stata data, saving ggplots
haven: read_dta()
library(haven)data <- read_dta("stata_data.dta")ggplot2: ggsave()
library(ggplot2)p <- forest_plot(ma, group = group)ggsave(p, "forest_plot.png", dpi = 320, height = 8)66 / 73
tidymeta
67 / 73
tidymeta
meta_analysis()/your_favorite_function() + tidy()
68 / 73
tidymeta
meta_analysis()/your_favorite_function() + tidy()
forest_plot()/text_table()
69 / 73
tidymeta
meta_analysis()/your_favorite_function() + tidy()
forest_plot()/text_table()
sensitivity()/influence_plot()
70 / 73
tidymeta
meta_analysis()/your_favorite_function() + tidy()
forest_plot()/text_table()
sensitivity()/influence_plot()
cumulative()/cumulative_plot()
71 / 73
Resources
R for Data Science: A comprehensive but friendly introduction to the tidyverse. Free online.
DataCamp: ggplot2 courses and tidyverse courses
ggplot2: Elegant Graphics for Data Analysis: The official ggplot2 book
72 / 73

github.com/malcolmbarrett/tidymeta
github.com/malcolmbarrett/ma_viz_workshop
Slides created via the R package xaringan.
73 / 73